Profiling Python
This article explains the basics of profiling Python code. The hardest part is installing all the great tools that make it trivial to find the bottleneck in your code.
Getting the required tools
This can be a pain in the neck on a Mac. The rule of thumb I adhere to is Use
conda, pip and homebrew where possible.
Python and core packages (numpy, scipy, etc)
Installing Python modules can sometimes be challenging as these link to compiled
C/Fortran code behind the scenes. I recommend using conda, which can
install pre-compiled modules for your operating system. Download from
http://conda.pydata.org/miniconda.html. conda can be installed to your home
directory, which comes in handy on the cluster.
After that, you can easily set up a virtual environment and install many of the core packages:
conda create -n py3k anaconda python=3
conda install numpy
The first step above should prepend the newly set up Python interpreter to your
path. If it doesn’t, prepend the path to the new interpreter by editing
~/.bashrc and ~/.bash_profile. Pip should also be installed— if it is not,
type the following into a terminal
conda install pip
Once conda and pip are up and running, you should always attempt to install
new Python packages through them instead of from source. This will save a lot of
headache in the long run as you won’t have to compile C/Fortran code.
Note
Some Python packages serve as a wrapper around other software, such as MySQLdb
around mysql and bsddb around BerkeleyDB. The underlying software is best
installed via homebrew. For instance, here is how to set up BerkeleyDB:
# install C/Fortran compilers, these will come in handy all the time
brew install gcc
# install berkeley to /usr/local/Cellar/berkeley-db/<VERSION>
brew install berkeley-db
# install python wrapper, passing in the path to the berkeley-db installation
BERKELEYDB_DIR=/usr/local/Cellar/berkeley-db/5.3.15/ pip install bsddb3
Profiling result visualisation (Runsnakerun/ Snakeviz)
These tool are useful for finding which function calls take the most time. You cannot look inside of functions using these. The next section will explain how to profile a single function to find which lines take the most time.
I know of two great packages the can visualise the output of the Python
profiler. snakeviz has some fancy features and written in pure Python. It is
easier to set up and use, but somewhat slow. If you have profiled a large chunk
of an application, snakeviz will give up. In practice, I found it’s only
useful for small programs. runsnakerun is written in C, so it can display
just about anything, but is somewhat trickier to install.
pip install snakeviz
pip install runsnakerun
For runsnakerun, you may also have to install SquareMap via brew/conda/pip.
To visualise the results of a profiling run with snakeviz type
snakeviz out.prof
where out.prof is the file output by the profiler (see below). If you get
“Sorry, we were not able to load this profile! You can try profiling a smaller
portion of your code.”, then you will have to use runsnakerun:
ipython qtconsole
In the window that opens, type
from runsnakerun import runsnake
runsnake.main()
then use the GUI to open out.prof.
The reason we are using ipython instead of launching runsnakerun from the
terminal is that OSX restricts some applications’ access to the screen. The
version of runsnakerun installed through pip is not an Apple “framework” and
therefore cannot access the screen. There are multiple guides explaining how to
get around this on the Internet, but none of them has worked for me.
IPython extensions
There are many great tutorials on how to set up and use the required IPython extensions, such as http://pynash.org/2013/03/06/timing-and-profiling.html .
These extensions are best used to profile single functions (see below). lprun
and mprun are of particular interest. Also note prun prints as a table the
same information that snakeviz/runsnakerun visualises. Personally I find
the visualisation much more useful.
Profiling the entire application
To profile an entire application, run
python -m cProfile -o out.prof mycode.py
This creates a file called out.prof, which can be visualised using snakeviz
or runsnakerun. The visualisation might like this:
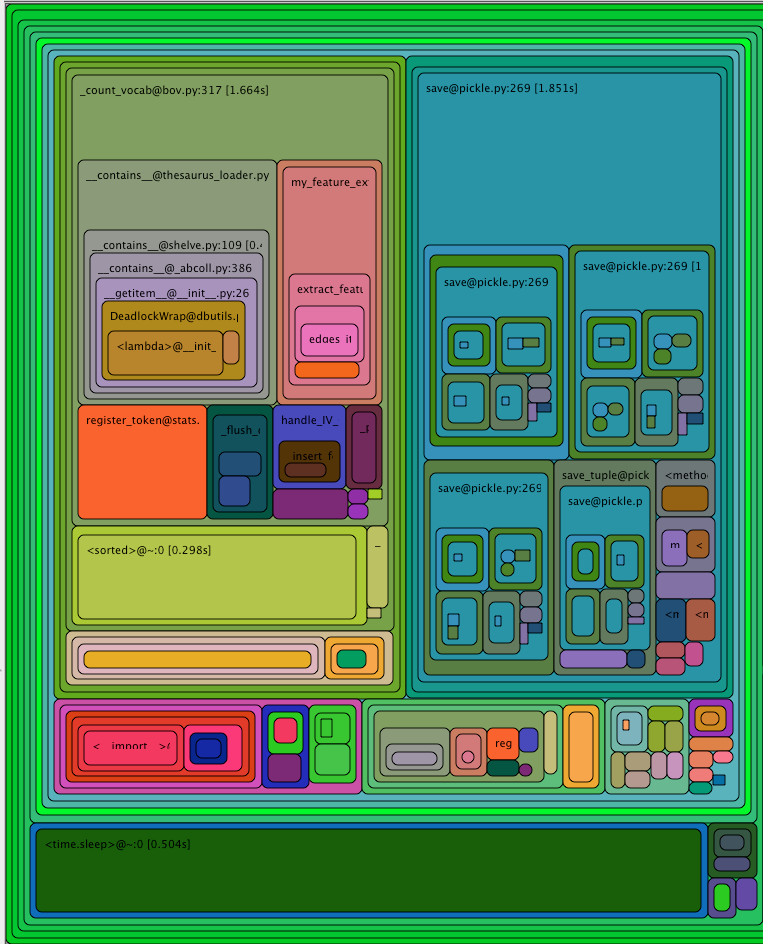
The top left corner of that image can be read as
- function
_count_vocabtook a total of 1.6s - 0.6s of which were spent in
__contains__(not visible in the image, you need to hover over the box inrunsnakerun) - 0.2s of which were spent in
__my_feature_extractor__, etc
This technique can be used to find out which functions take a long time.
Profiling functions
Having set up the required IPython extensions and identified bottleneck functions, we can profile and incrementally optimise these. Let’s illustrate this with a real example I was recently working on.
I have a function called calculate_log_odds, which does something rather
simple but seems to take a lot of time. To profile it line by line, I ran the
following command in IPython
# get some data to run the function with
import pickle
b = pickle.load(open('../statistics/stats-exp0-0-cv0-ev.MultinomialNB.pkl'))
X = b.tr_matrix
y = b.y_tr
# do the actual profiling
%lprun -f calculate_log_odds calculate_log_odds(X, y)
The output looks like this:
Total time: 2.36018 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
3 def calculate_log_odds(X, y):
4 # todo this is quite slow
5 1 14 14.0 0.0 log_odds = np.empty(X.shape[1])
6 1 45 45.0 0.0 class0_indices = (y == sorted(set(y))[0])
7 2466 1632 0.7 0.1 for idx in range(X.shape[1]):
8 2465 864021 350.5 36.6 all_counts = X[:, idx].A.ravel() # document counts of this feature
9 2465 1378602 559.3 58.4 total_counts = sum(all_counts > 0) # how many docs the feature occurs in
10 2465 79524 32.3 3.4 count_in_class0 = sum(all_counts[class0_indices]) # how many of them are class 0
11 2465 3871 1.6 0.2 p = float(count_in_class0) / total_counts;
12 2465 29874 12.1 1.3 log_odds_this_feature = np.log(p) - np.log(1 - p)
13 2465 2595 1.1 0.1 log_odds[idx] = log_odds_this_feature
14 1 0 0.0 0.0 return log_odds
Most of the time is spent on line 9, where I am using the Python sum function
to add up a numpy array. This can be made much faster by using the appropriate
numpy function:
Function: calculate_log_odds2 at line 16
Total time: 1.54092 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
16 def calculate_log_odds2(X, y):
17 1 16 16.0 0.0 log_odds = np.empty(X.shape[1])
18 1 55 55.0 0.0 class0_indices = (y == sorted(set(y))[0])
19 1 526 526.0 0.0 X = X.tocsc()
20 2466 1746 0.7 0.1 for idx in range(X.shape[1]):
21 2465 1417910 575.2 92.0 all_counts = X[:, idx].A.ravel() # document counts of this feature
22 2465 49792 20.2 3.2 total_counts = np.sum(all_counts > 0) # how many docs the feature occurs in
23 2465 32936 13.4 2.1 count_in_class0 = np.sum(all_counts[class0_indices]) # how many of them are class 0
24 2465 4377 1.8 0.3 p = float(count_in_class0) / total_counts;
25 2465 30788 12.5 2.0 log_odds_this_feature = np.log(p) - np.log(1 - p)
26 2465 2769 1.1 0.2 log_odds[idx] = log_odds_this_feature
27 1 1 1.0 0.0 return log_odds
Note the running time has gone down from 2.3s to 1.5s. Now the bottleneck of the function seems to be at line 21, where I slice a sparse matrix and unnecessarily convert to it a dense one. After a few more iterations of 1) find bottleneck line, and 2) correct line, I arrived at the following:
Total time: 0.058181 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
56 def calculate_log_odds5(X, y):
57 1 13 13.0 0.0 log_odds = np.empty(X.shape[1])
58 1 47 47.0 0.1 class0_indices = y == sorted(set(y))[0]
59 1 619 619.0 1.1 X = X.A
60 2466 1792 0.7 3.1 for idx in range(X.shape[1]):
61 2465 8277 3.4 14.2 all_counts = X[:, idx] # document counts of this feature
62 2465 7354 3.0 12.6 total_counts = np.count_nonzero(all_counts) # how many docs the feature occurs in
63 2465 8044 3.3 13.8 count_in_class0 = np.count_nonzero(all_counts[class0_indices]) # how many of them are class 0
64 2465 3521 1.4 6.1 p = float(count_in_class0) / total_counts
65 2465 25890 10.5 44.5 log_odds_this_feature = np.log(p) - np.log(1 - p)
66 2465 2623 1.1 4.5 log_odds[idx] = log_odds_this_feature
67 1 1 1.0 0.0 return log_odds
The running time is now 0.05 seconds, or a speedup of ~50x. The bottleneck
operation is now entirely done on numpy arrays, so there are no more
low-hanging fruit.
Notes
- Having unit tests for the functions that you are optimising is essential to making sure nothing gets broken.
- Functions to be profiled line by line cannot be typed into the interactive shell and must be imported from a file on disk